PROJECT LEADERS
Derek Ng, Assistant Professor, Department of Biology, University of Toronto Mississauga; Master of Science in Biomedical Communications Program, Institute of Medical Science, Temerty Faculty of Medicine, University of Toronto.
Stavroula Andreopoulos, Professor, Teaching Stream, Department of Biochemistry, Temerty Faculty Medicine, University of Toronto.
In each of the following sections, you will find a brief project description, links to the visual and interactive media, associated publications, funding sources and credits.
ENZYME KINETICS
Project Description
Michaelis-Menten enzyme kinetics is a major topic taught in undergraduate introductory biochemistry courses that serves as a foundation for other areas in the life sciences such as pharmacology and molecular biology. Key kinetic concepts and behaviours are typically modeled and explained using simplified reaction schematics, equations, and graphs. However, these abstract mathematical/symbolic representations often make it difficult for life science students to conceptually link pertinent concepts to the underlying molecular phenomena being described. In this project, we aim to:
- Design and develop both animated and interactive media that utilizes different co-visualization and interactive strategies to help students contextualize and relate the mathematical/graphical/abstract models of kinetic phenomena to the underlying molecular system being described.
- Evaluate whether these strategies/media help students develop a deeper molecular understanding of enzyme kinetics compared to more traditional teaching approaches.
Links to Educational Media
3D Animation

This 3D animation uses both realistic and metaphoric depictions of the underlying molecular players, environments, and interactions in Michaelis-Menten enzyme kinetics to contextualize and explain the relationship between the mathematical models and underlying molecular systems.
Interactive eModule
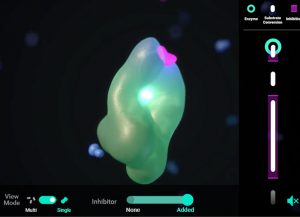
This eModule is a web‐based multimedia interactive learning tool designed to help students better bridge the relationships between these abstract mathematical models and the molecular behaviors, interactions, and dynamics that produce kinetic phenomena. It incorporates simulations, interactive leaflets, knowledge checks, and comparative summaries.
Publications
- U. Florjanczyk, J. Gu, S. Andreopoulos, and D.P. Ng. Enzyme Kinetic Digital Learning Tools: Their Use in Undergraduate Education. In preparation – to be submitted to Biochemistry and Molecular Biology Education, 2021
- J. Gu, S. Andreopoulos, J. Jenkinson, and D.P. Ng. 2020. Rethinking enzyme kinetics: designing and developing a biomolecular interactive tutorial (BIOMINT) learning tool for undergraduate students. Biochemistry and Molecular Biology Education. 48(1): 74-9.
- U. Florjanczyk, D.P. Ng, S. Andreopoulos, and J. Jenkinson. 2018. Developing a 3D animation for deeper molecular understanding of Michaelis-Menten enzyme kinetics. Biochemistry and Molecular Biology Education. 46: 561-5.
Presentations, Abstracts & Posters
- U. Florjanczyk, Z. Gu, S. Long, C. Tang Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Engaging biochemistry students with storytelling, simulations, and gaming. Poster presentation at the 2020 Biennial Conference on Chemical Education. Abstract accepted on March 31, 2020. Conference canceled due to COVID.
- U. Florjanczyk, Z. Gu, S. Long, C. Tang Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Stories, simulations, and games: designing technologically-enhanced educational tools to support and engage biochemistry students. Poster presentation at the 2020 Biennial Conference on Chemical Education. Abstract accepted on March 31, 2020. Conference canceled due to COVID.
- J. Gu, S. Andreopoulos, J. Jenkinson, and D.P. Ng. BIOMINT: a digital interactive tutorial to facilitate deeper molecular understanding of Michaelis-Menten enzyme kinetics. Poster presentation at the McMaster Research on Teaching and Learning Conference. McMaster University, Hamilton, Ontario. December, 2018.
- D. P. Ng and S. Andreopoulos. Visual and interactive media for deeper molecular understanding of enzyme kinetics. Oral presentation at the Arts & Science Teaching and Learning Projects Showcase, 2nd Annual Meeting. Faculty of Arts & Science, University of Toronto, Toronto, Ontario. April, 2018.
- U. Florjanczyk, D.P. Ng, S. Andreopoulos, and J. Jenkinson. Developing a 3D animation for deeper molecular understanding of Michaelis-Menten enzyme kinetics. Poster presentation at the Western Conference on Science Education. Western University, London, Ontario. July, 2017.
Funding Sources
- D. P. Ng and S. Andreopoulos. Visual and interactive media for deeper molecular understanding of enzyme kinetics. Advancing Teaching and Learning in Arts & Sciences (ATLAS). $15,000, 2017 – 2019.
Credits
- 3D animation – Ursula Florjanczyk, Graduate Student, Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
- Interactive eModule – Jerry Gu, Graduate Student, Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
- Pedagogical Evaluation – Jodie Jenkinson, Associate Professor, Department of Biology, University of Toronto Mississauga; Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
METABOLISM
Project Description
Metabolism, including cellular respiration, encompasses a large component of the topics covered in foundational biochemistry and life science undergraduate courses. The difficulty in instruction and student retention of this topic lies in the conceptualization of the numerous enzymatic reactions involving various substrates/products and their regulation. Learners typically end up focusing on memorizing individual metabolic pathways and their subcomponents (e.g., enzymes, substrates, and products) rather than developing a high-level understanding of the interplay and regulation occurring between pathways and appreciating the real-life application of this knowledge. The aims of this project are to:
- Design and evaluate digital teaching tools that balance the sheer volume of information (e.g., enzymatic reactions, competing pathways, and points of regulation) with a higher level understanding of energy needs and how the metabolic flux of the cell adapts to these changing needs.
Links to Educational Media
3D Animation
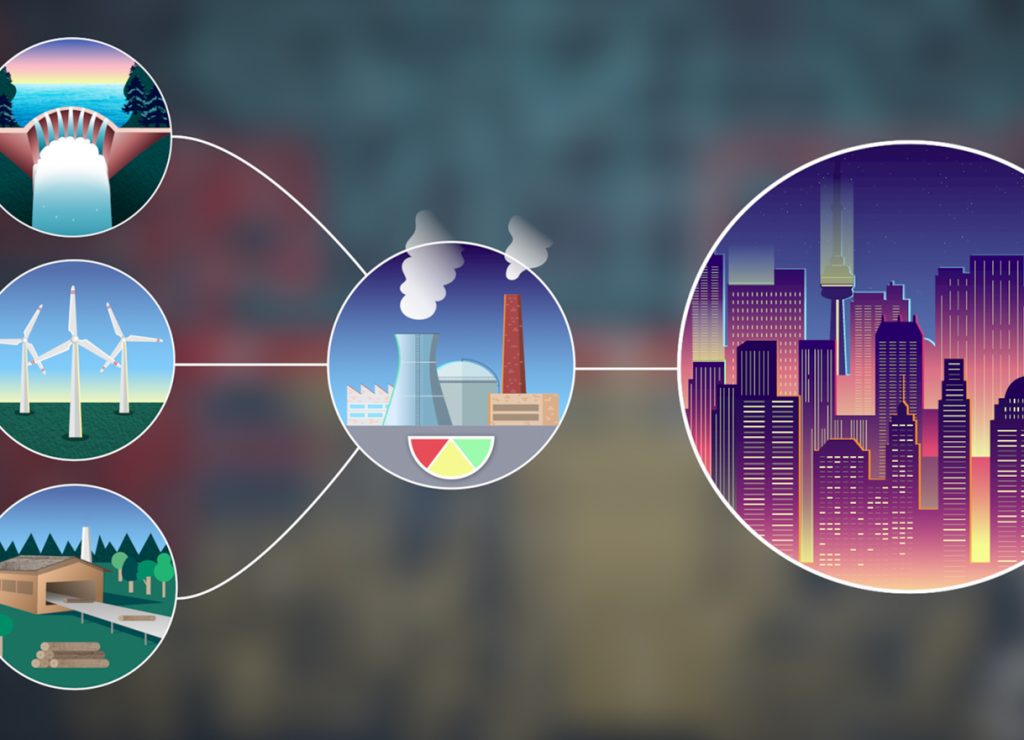
This 3D animation introduces students to the fundamental concepts of glucose and fat metabolism, energy production by the citric acid cycle (CAC) and oxidative phosphorylation, as well as the major points of regulation that emphasizes the integration and flux of these metabolic pathways.
Digital Game
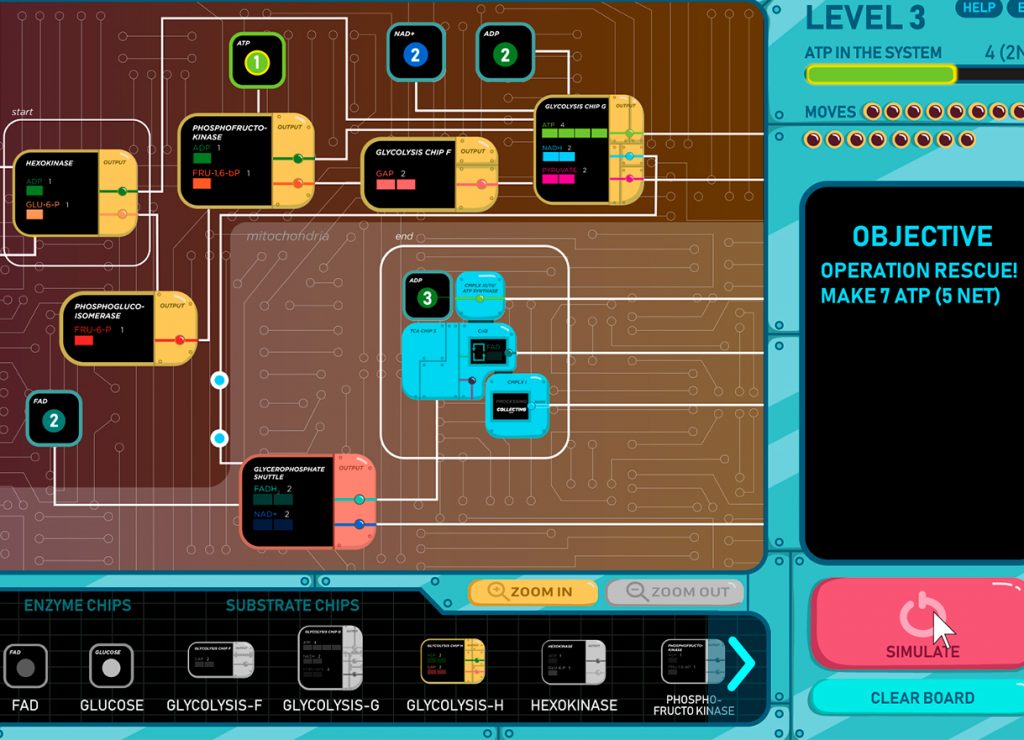
This video is a conceptual demo showing level one of an eight-level 2D game, “Sugar Scramble”. This game uses a circuit board metaphor representing the metabolic system. In order to win each level, circuit board connections need to be made between enzymes and pathways to generate a certain target amount of energy. Our pedagogical goal for this game is to allow the players to interactively apply and integrate their understanding of metabolic flux and to visualize how it can change depending on energy demands.
Publications
- C. T. Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Sugar scramble: a game based approach to learning glucose metabolism. Work in progress, 2021.
- C. T. Poy, S. Andreopoulos, S. Patterson, J. Jenkinson and D. P. Ng. Learning Glucose Metabolism Through “Sugar Scramble”: A digital Game-Based Approach. 15th European Conference on Game Based Learning (ECGBL), Conference Proceedings, Brighton, UK, Sept 22, 2021.
- S. Long, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Metabolism in motion: engaging biochemistry students with storytelling. Journal of Chemical Education. 98 (5): 1795-1800, May 2021 (plus May 2021 Journal Cover Art)
- D. P. Ng, S. Andreopoulos, S. Long, C. Tang Poy, S. Patterson, and J. Jenkinson. 2020. Enhancing learning of glucose metabolism through animations and gaming. The 6th E-Learning Excellence Awards: An Anthology of Case Histories, edited by Dan Remenyi, Academic Conferences International Limited. 165–75.
Presentations, Abstracts, & Posters
- C. T. Poy, S. Andreopoulos, S. Patterson, J. Jenkinson and D. P. Ng. Learning Glucose Metabolism Through “Sugar Scramble”: A digital Game-Based Approach. 15th European Conference on Game Based Learning (ECGBL), Conference Proceedings, Brighton, UK, Sept 22, 2021.
- S. Long, C. Tang Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, D.P. Ng. Enhancing learning of glucose metabolism through animations and gaming. Oral presentation for The 6th E-Learning Excellence Awards at the 19th European Conference on e-Learning. Virtual, University of Applied Sciences HTW, Berlin, Germany. Oct. 28, 2020.
- C. T. Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Sugar scramble: a game-based approach to learning glucose metabolism. Poster presentation at the 19th European Conference on e-Learning. Virtual, University of Applied Sciences HTW, Berlin, Germany. Oct. 30, 2020.
- S. Long, C. Tang Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, D.P. Ng. Metabolism in motion: engaging biochemistry students with storytelling and gaming. Poster presentation at the 13th International Conference on e-Learning & Innovative Pedagogies. Virtual, University of the Aegean, Rhodes, Greece. April 24, 2020.
- S. Long, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. Metabolism in motion: engaging biochemistry students with storytelling. Oral presentation at the Association of Medical Illustrators Annual Meeting. Milwaukee, Wisconsin. July, 2019.
- C. T. Poy, S. Andreopoulos, S. Patterson, J. Jenkinson, and D.P. Ng. A digital game-based approach to learning glucose metabolism. Oral presentation at the Association of Medical Illustrators Annual Meeting. Milwaukee, Wisconsin. July, 2019.
Funding Sources
- D. P. Ng, S. Andreopoulos, and S. Patterson. Sugar Scramble: A Game-Based Approach to Learning Glucose Metabolism. Instructional Technology Innovation Fund (ITIF). $5,000, 2021 – 2022.
- D. P. Ng, S. Andreopoulos, and S. Patterson. Enhancing learning of glucose metabolism through animated and interactive media. Advancing Teaching and Learning in Arts & Sciences (ATLAS). $15,000, 2018 – 2020.
Credits
- 3D animation – Shirley Long, Graduate Student, Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
- Sugar Scramble Game – Colleen Tang Poy, Graduate Student, Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
- Subject Matter Expert – Sian Patterson, Associate Professor, Teaching Stream, Department of Biochemistry, Temerty Faculty of Medicine, University of Toronto
- Pedagogical Evaluation – Jodie Jenkinson, Associate Professor, Department of Biology, University of Toronto Mississauga; Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
ALTERNATIVE ROLES OF RIBOSOMAL PROTEINS
Project Description
Ribosomes are complex molecular machines that directly translate the genetic information encoded by messenger RNA (mRNA) into the twenty amino acid code for proteins. The structure of the ribosome contains both ribosomal RNA (rRNA) and ribosomal proteins where the latter stabilizes the overall ribosomal structure. Recent research indicates that ribosomal proteins may have roles beyond ribosome structural scaffolding. The aims of this project are to:
- Facilitate student visualization and understanding of the extra-ribosomal roles of ribosomal proteins using a visual storytelling approach.
- Accurately visualize 3D structures of ribosomes, ribosomal proteins and other molecules that are involved in extra-ribosomal activity of ribosomal proteins.
- Serve as a stepping stone that assists students in navigating the complex and rapidly expanding field of ribosome-related research.
Links to Educational Media
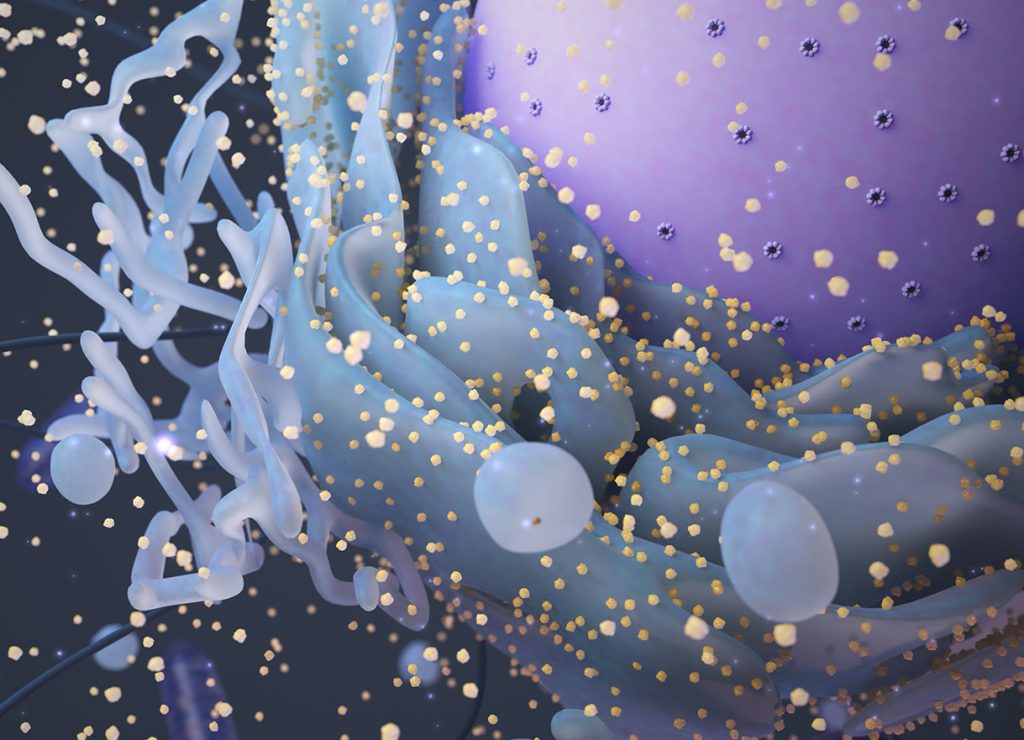
3D Animation
This 3D animation highlights how the molecular structure and interactions of certain ribosomal proteins permit them to serve dual roles as both structural scaffolds and cell-cycle regulators. Specifically, we highlight the altered degradation of p53 tumour suppressor protein by the direct interaction of mouse double minute 2 (MDM2) with ribosomal proteins. In addition, we touch upon the clinical consequences of improper ribosomal assembly leading to an imbalance between the dual roles for these ribosomal proteins.
Publications
- Z. Bai, S. Andreopoulos, and D.P. Ng. A 3D animation on the extra-ribosomal functions of ribosomal proteins. Work in progress, 2021.
Presentations, Abstracts, & Posters
- Z. Bai, S. Andreopoulos, and D.P. Ng. A 3D animation on the extra-ribosomal functions of ribosomal proteins. Poster presentation at the 14th International Conference on e-Learning & Innovative Pedagogies. Virtual, University of Aegean, Rhodes, Greece. Abstract accepted on May 15, 2020.
Credits
- 3D animation – Jenny Bai, Graduate Student, Master of Science in Biomedical Communications Program, Institute of Medical Science, University of Toronto.
CRISPR
Coming soon.
CONTACT US
We would appreciate your feedback on these educational tools. In particular, please share with us how you used them and what you thought of them.

